The exome includes all protein-coding regions, also known as exons, in the human genome. Although exons only comprise 1%–2% of the genome, approximately 89% of all known disease-causing mutations are in these regions. However, disease-relevant variants can also be found in non-coding regions. We developed ExomeXtra® Sequencing to also target disease-relevant variants in non-coding regions described in the Human Gene Mutation Database (HGMD) and ClinVar database.
In addition to these non-coding variants, we included a new and unique CNV backbone that enables genome-wide analysis and high-resolution detection of copy number variations (CNVs). This new CNV backbone allows CNV calling in coding regions and CNV detection in non-coding regions to increase the probability of finding disease-causing mutations.
Thus, our ExomeXtra® Sequencing products combine the advantages of whole exome sequencing (WES) and whole genome sequencing (WGS), covering all disease-relevant genomic regions.
CeGaT Is the Best Partner for Sequencing Your Project
Our Commitment to You
Fast Processing
Turnaround time
≤ 15 business days
High Quality
Highest accuracy for all processes
Secure Delivery
Secure provision of sequenced data via in-house servers
Safe Storage
Safe storage of samples and data after project completion
Our Service
We provide a comprehensive and first-class project support – from selecting the appropriate product to evaluating the data. Each project is supervised by a committed scientist. You will have a contact person throughout the whole project.
Our service includes:
- detailed project consulting
- product selection tailored to your project
- detailed bioinformatic evaluation of your data
- detailed project report with information about sample quality, sequencing parameters, bioinformatic analysis, and results
Benefit from our dedicated support and accredited workflows.
Explore Our Product Portfolio for ExomeXtra® Sequencing
We offer different ExomeXtra® Sequencing (EXS) products to address a variety of research questions. Would you like to have bioinformatic analyses performed on your data in addition to the included deliverables? Each of our products can be supplemented with further services. We are happy to advise you.
EXS Classic | EXS Premium | EXS Premium Deep |
Species | Species Human | Species Human |
DNA quality High molecular weight DNA | DNA quality Various/poor quality (e.g., fragmented DNA, low input) | DNA quality Various/poor quality (e.g., fragmented DNA, low input) |
Target enrichment ExomeXtra® Sequencing | Target enrichment ExomeXtra® Sequencing | Target enrichment ExomeXtra® Sequencing |
Targeted region Whole exomic regions | Targeted region Whole exomic regions | Targeted region Whole exomic regions |
Sequencing platform Illumina | Sequencing platform Illumina | Sequencing platform Illumina |
Output 18 Gb | Output 18 Gb | Output 24 Gb |
Included deliverables Project report & files in FASTQ format | Included deliverables Project report & files in FASTQ format | Included deliverables Project report & files in FASTQ format |
Bioinformatics
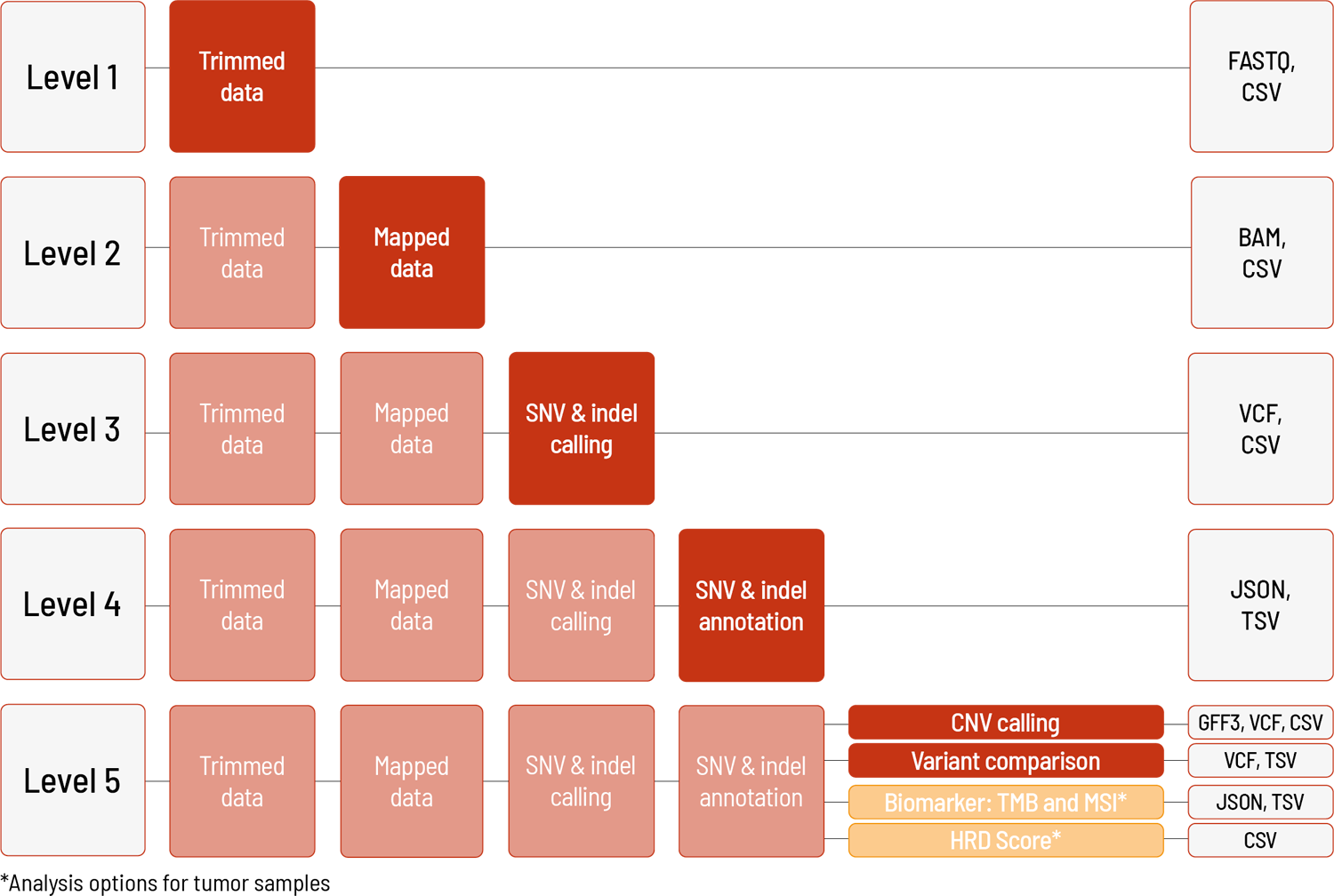
Raw sequencing data are automatically processed. We offer different levels of bioinformatic analysis. The default level is Level 1. With increasing bioinformatic level, more data are delivered. All higher levels include the data from the lower levels. In addition to the data, and independent of the analysis level, a project report is generated.
All EXS products are analyzed with the Illumina DRAGEN Bio-IT Platform. We can perform the analyses based on the human references hg19 or GRCh38.
Level 1:
- demultiplexing and adapter trimming of the sequencing data (FASTQ format)
- metrices (CSV format)
- MultiQC report (HTML format)
Level 2:
- mapping of the sequencing data (BAM format)
- metrices (CSV format)
Level 3:
- calling of single nucleotide variants (SNVs) and small insertions and deletions (indels) (VCF format)
- metrices (CSV format)
Level 4:
- annotation of the SNVs and indels (JSON and TSV format) (only for human samples)
Level 5 (one of the following):
- calling of copy number variations (CNVs) (VCF and GFF3 format), including metrices (CSV format)
- variant comparison in multiple samples, e.g., trio filtering (VCF and TSV format)
For tumor samples (tumor-only and tumor-normal comparison), we can additionally offer the following analysis:
- biomarker determination: TMB and MSI (TSV and JSON format)
For tumor samples with corresponding normal tissue (tumor-normal comparison), we can additionally offer the following analysis:
- calculation of HRD score (CSV format)
Technical Information
At CeGaT, paired-end sequencing (2 x 100 bp) is performed using the Illumina sequencing platforms.
Please be aware that our ExomeXtra® Sequencing is updated regularly for constant improvement. During these updates, diagnostically relevant targets are added or removed based on the latest scientific knowledge. Please be aware that we can only offer the newest version. Thus, no previous versions can be requested.
Further Information about ExomeXtra® Sequencing
The exome harbors 89% of all disease-causing mutations, while it only accounts for 1%-2% of the genome. However, additional genetic information might be required to solve complex disease patterns. Off-the-shelf exomes often miss disease-relevant variants in intronic or regulatory regions. Therefore, we designed the ExomeXtra® to include all known pathogenic regions in the human genome – independent of their location in introns or exons.
In the first designing step, we evaluated available technologies to find the one that gives the most complete and uniform coverage of all disease-causing targets. These targets may not only be located in coding but also in non-coding regions of the genome. Based on these findings, we designed the ExomeXtra® enrichment with the best coverage of all known disease-causing variants. After sequencing our ExomeXtra®, the high-quality data is processed by our bioinformatics pipeline with multiple options for bioinformatic analyses.
Our ExomeXtra® Sequencing products are updated regularly with the latest scientific knowledge. During these updates, new non-coding variants from relevant databases, such as HGMD or ClinVar, that are classified as pathogenic or likely pathogenic are included in our ExomeXtra® enrichment.
Simultaneously, variants no longer classified as pathogenic or likely pathogenic are removed. Thus, our ExomeXtra® Sequencing products include all genome regions described as disease-relevant in the respective databases.
With our ExomeXtra® Sequencing, we can even detect deletions and duplications in the entire genome. The new and unique CNV backbone enables genome-wide analysis and the detection of copy number variations (CNVs) at a high resolution. The CNV detection in both coding and non-coding regions is unique in exome sequencing. Our ExomeXtra® Sequencing provides uniform coverage and an increasing probability of finding disease-causing mutations.
Our in-house design of ExomeXtra® incorporates clinically relevant sections of non-coding RNA genes, flanking intronic sequences, and the entire mitochondrial genome. Thus, ExomeXtra® Sequencing is a good approach to finding the genetic cause of a disease.
Downloads
Contact Us
Do you have a question or are you interested in our service? Feel free to contact us. We will take care of your request as soon as possible.
Start Your Project with Us
We are happy to discuss sequencing options and to find a solution specifically tailored to your clinical study or research project.
When getting in contact, please specify sample information including starting material, number of samples, preferred library preparation option, preferred sequencing depth and required bioinformatic analysis level, if possible.