Gene fusions are often drivers of cancer. Therefore, drugs targeting these gene fusions are increasingly used in treating tumor patients.
CeGaT has developed CancerFusionRx®, a unique method to detect structural changes (gene fusions) at the RNA level. This method is far superior to DNA-based methods and transcriptome-level approaches and allows the detection of all known as well as novel gene fusions. Based on the gene fusions found, we can provide specific drug recommendations. CancerFusionRx® is thus a valuable tool in genetic tumor diagnostics and supports you in choosing the best possible therapy.
Are you insured in Germany? Our colleagues at the Zentrum für Humangenetik Tübingen will gladly support you!
What We Offer with This Service
Our Promise to You
Service Details
- Over 200 genes for fusion detection and over 120 exon-exon-specific enrichments targeting recurrent breakpoints
- A list of all eligible drugs, with EMA and/or FDA approval, for which corresponding gene fusions could be detected in the tumor — learn more
We will report all relevant findings in a medical report. This contains a list of all identified clinically relevant structural alterations. The fusions are annotated with gene name, functional class, transcript-ID, and effect on protein function. Each single medical report is prepared and discussed by an interdisciplinary team of scientists and physicians to guarantee highest quality.
Method
RNA isolated from tumor material is transcribed into cDNA and prepared for sequencing on the NovaSeq™ 6000 using an optimized enrichment protocol. The bait design includes the possibility of targeted detection of known relevant gene fusions as well as the detection of not yet described gene fusions as one fusion is part of the enrichment. In addition, intragenic transcript variants such as MET exon 14 skipping and activating EGFR deletions with therapeutic relevance can be detected.
Following data processing, our team of scientists and specialists in human genetics analyze the data and issue a medical report.
Sample Report
General Information
Our Standard Sample Requirements
Sample requirements for all analyses (minimum 20% tumor content):
- FFPE tumor block (recommended sample type) or
- FFPE tissue slides (minimum 10 slides).
- If possible: H&E-stained slides with tumor area distinctly labeled. Please report the tumor content (of the labeled tumor area).
Here you can find more information on how to ship your sample safely.
Turnaround Time
- Turnaround Time: 2-3 Weeks
Please feel free to contact us via contact form or write us an email at tumor@cegat.de for further details.
This Is What Makes Our CancerFusionRx® Service Special
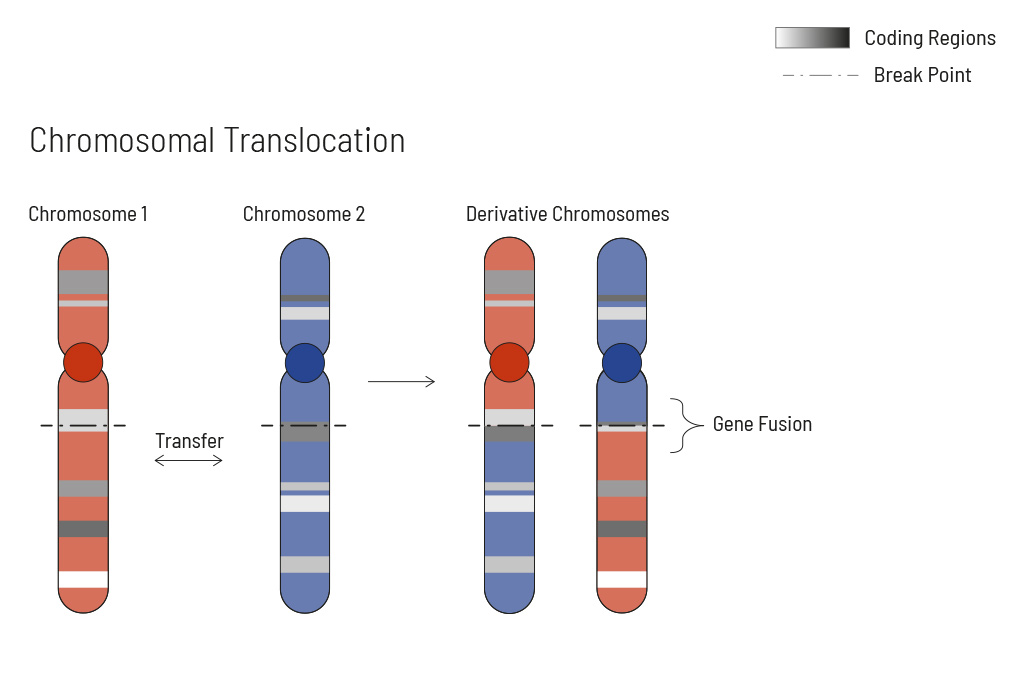
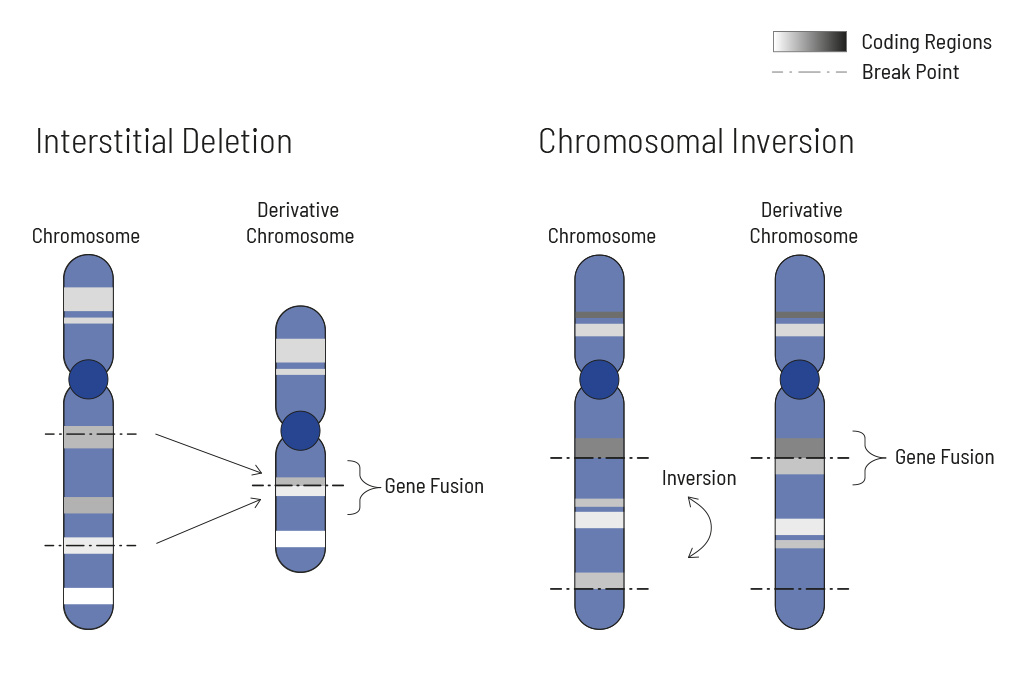
Chromosomal rearrangements frequently occur in all types of cancer. As a result, gene fusions can occur in the cancer genome. Fusions are major drivers of cancer and are therefore most relevant for treatment decisions. Conventional PCR-based methods will not detect a fusion when the other partner is not known (frequently relevant for neutrophic tyrosine kinase, NTRK fusions). Even whole transcriptome analyses are not sensitive enough, especially when the tumor content is low.
To detect all known and previously described as well as novel gene fusions with a therapeutic option, we developed a next-generation targeted enrichment on RNA-basis. The design currently includes over 200 genes for fusion detection and over 120 exon-exon-specific enrichments with known breakpoints. This method is superior to DNA-based methods and also to whole RNA-based approaches. We strongly recommend completing the genetic tumor diagnostic by RNA enrichment for fusions for the most complete understanding of the tumor’s biology.
Structural Variants with Potential Therapeutic Relevance
Guidance on potentially effective drugs
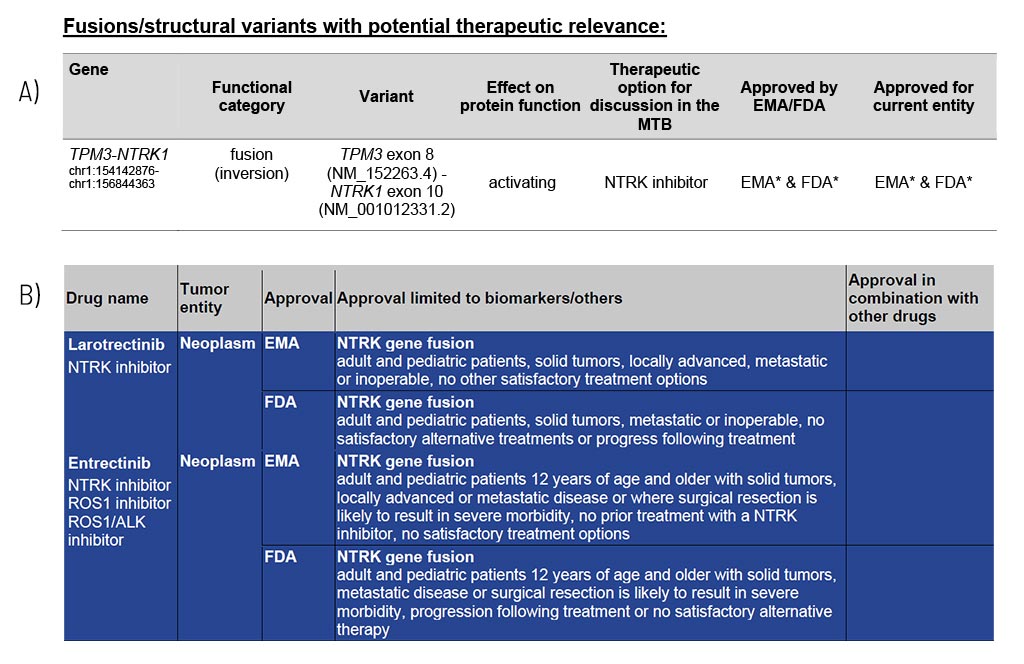
The identified fusions are shown in detail, along with resulting information on the potential therapy options, including EMA/FDA approval (A). These therapy options are the foundation for the specialists of a molecular tumor board (MTB) to discuss an individual treatment recommendation for each patient.
In the appendix/supplement of the report, we list relevant therapeutics for each identified structural variant (B). This compilation provides you with information about drug classes and names, their approval (FDA/EMA), and associated restrictions.
Sample Report: Exemplary for the TPM3-NTRK1 fusion detected and the resulting therapeutic options in a colorectal cancer patient. Top panel (A): An excerpt from Table 1 of the findings, listing fusions/structural variants with therapeutic relevance. Lower panel (B): Listing and description of the drugs.
Gene Directory
Gene list for RNA-based identification of fusion transcripts (CancerFusionRx®, STR01)
Gene list for de-novo fusion detection
ABL1, ACTB, AFAP1, AGK, AKAP4, AKAP9, AKAP12, AKT1, AKT2, AKT3, ALK, ARHGAP6, ARHGAP26, ASPL, ASPSCR1, ATF1, ATP1B1, ATRX, AVIL, AXL, BAG4, BCL2, BCOR, BCORL1, BCR, BEND2, BICC1, BRAF, BRD3, BRD4, c11orf95, CAMTA1, CCAR2, CCDC6, CCDC88A, CCDC170, CCNB3, CCND1, CD44, CD74, CEP85L, CIC, CLDN18, CLIP1, CLTC, CNTRL, COL1A1, CREB1, CREB3L1, CREB3L2, CRTC1, CTNNB1, DDIT3, DNAJB1, EGFR, EML4, EPC1, EPCAM, ERBB2, ERBB4, ERG, ESR1, ESRRA, ETV1, ETV4, ETV5, ETV6, EWSR1, EZR, FEV, FGFR1, FGFR2, FGFR3, FLI1, FN1, FOXO1, FOXO4, FOXR2, FUS, GLI1, GOPC, GPR128, HEY1, HMGA2, HTRA1, IGF1R, INSR, JAK2, JAZF1, KIAA1549, KIF5B, KIT, LEUTX, LMNA, LPP, LTK, MAGI3, MAML1, MAML2, MAML3, MAMLD1, MAP3K8, MARS1, MAST1, MAST2, MEAF6, MET, MGA, MGMT, MITF, MKL2, MN1, MSH2, MYB, MYBL1, MYC, NAB2, NCOA1, NCOA2, NCOA3, NCOA4, NFATC2, NFIB, NOTCH2, NPM1, NR4A3, NRG1, NRG2, NSD3, NTRK1, NTRK2, NTRK3, NUTM1, PAX3, PAX7, PAX8, PBX1, PDGFB, PDGFD, PDGFRA, PDGFRB, PHF1, PIK3CA, PLAG1, PML, POU5F1, PPARG, PPARGC1A, PPP1CB, PRKACA, PRKAR1A, PRKCA, PRKCB, PRKD1, PRKD2, PRKD3, PTPRZ1, QKI, RAD51B, RAF1, RANBP2, RARA, RELA, RELCH, RET, ROS1, RPS6KB1, RREB1, RSPO2, RSPO3, SDC1, SDC4, SH3PXD2A,SLC1A2, SHTN1, SLC34A2, SLC44A1, SLC45A3, SND1, SQSTM1, SS18, SSX1, SSX2, SSX4, STAT6, STRN, SUZ12, TACC1, TACC2, TACC3, TAF2N, TAF15, TCF3, TCF12, TERT, TFE3, TFEB, TFG, THADA, TMPRSS2, TPM3, TPR, TRIM24, TRIM33, TRIO, TTYH1, VGLL2, VGLL3, VMP1, WT1, WWTR1, YAP1, YWHAE, ZC3H7B, ZMYM2, ZNF703
Gene list for selected break points in these fusion genes
AFAP1-NTRK2, ATP1B1-NRG1, BCOR-CCNB3, BRD3-NUTM1, BRD4-NUTM1, CCDC6–RET, CCDC88A-ALK, CD74-NRG1, CD74-ROS1, CLTC-ALK, DNAJB1-PRKACA, EGFR-PPARGC1A, EML4-ALK, ETV6 NTRK2, ETV6-NTRK3, EWSR1-ATF1, EWSR1-ERG, EWSR1-FLI1, EWSR1-WT1, EZR-ROS1, FGFR1-TACC1, FGFR2-BICC1, FGFR2 TACC3, FGFR3-TACC3, KIAA1549-BRAF, KIF5B-ALK, KIF5B–RET, MGA-NUTM1, NAB2-STAT6, NCOA4–RET, NPM1-ALK, NSD3-NUTM1, PAX3-FOXO1, PAX7-FOXO1, PPP1CB-ALK, PRKAR1A-RET, QKI-NTRK2, RANBP2- ALK, RPS6KB1–VMP1, SDC4-NRG1, SDC4 ROS1, SLC34A2-ROS1, SND1-BRAF, SS18-SSX1, SS18-SSX2, STRN ALK, TMPRSS2-ERG, TPM3-ALK, TPM3-NTRK1, TPM3-ROS1, TPR NTRK1, TRIM24-BRAF, TRIM24-NTRK2, TRIM33-RET, TRIO-TERT
List for specific transcript variants
EGFR del ex2-22 (mLEEK), EGFR del ex25-26 (EGFRvIVb), EGFR del ex25-27 (EGFRvIVa), EGFR del ex26-27, EGFR del ex14-15 (vII), EGFR del ex2-7 (vIII), FGFR2IIIb, MET ex14 skipping, NFE2L2 ex2 skipping, PDGFRA del ex8-9
Downloads
Download Order Form
(requestable via CancerPrecision®, page 3)
Contact Us
Do you have a question, or are you interested in our service?
Diagnostic Support
We will assist you in selecting the diagnostic strategy – for each patient.