CancerEssential® has been developed in cooperation with oncologists and pathologists in order to provide a fast, cost-efficient and high-quality analysis of the most important driver mutations within the most common tumor entities. The detection of these specific mutations in tumor tissue has a clear treatment consequence with respect to approved and targeted treatment.
Are you insured in Germany? Our colleagues at the Zentrum für Humangenetik Tübingen will gladly support you!
What We Offer with This Service
* Based on a high-quality sample with 20% tumor content for detecting a somatic heterozygous variant.
Our Promise to You
Your Benefits
- Multigene panel testing
- Analysis of single nucleotide variants (SNVs), selected fusion genes, and microsatellite instability (MSI) status
- High average sequencing coverage: 1,000x
- Sensitivity*: > 99.8%; Specificity: > 99.9%
- A list of all eligible drugs, with EMA and/or FDA approval, for which corresponding biomarkers could be detected in the tumor – learn more
We will report all relevant findings in a medical report. This includes a list of all identified clinical relevant variants. These variants are clearly annotated with gene name, functional category of the mutation, transcript-ID, allele frequency, and effect on protein function. If requested, we also report the microsatellite instability status. Each single medical report is prepared and discussed by an interdisciplinary team of scientists and physicians to guarantee highest quality.
* Based on a high-quality sample with 20% tumor content for detecting a somatic heterozygous variant.
Method
The enrichment of the coding regions and the adjacent intronic regions is performed using a in-solution hybridization technology. The selection of the targeted regions and the design of the enrichment baits is performed in-house. High-throughput sequencing is performed on our Illumina platforms. Bioinformatic processing of the data is achieved using an in-house computer cluster.
Following data processing, our team of scientists and specialists in human genetics analyze the data and issue a medical report.
Sample Report
General Information
Our Standard Sample Requirements
Sample requirements for all analyses (minimum 20% tumor content):
- DNA (> 200 ng) or
- FFPE tumor block (recommended sample type) or
- FFPE tissue slides (minimum 10 slides)
- If possible: H&E-stained slides with tumor area distinctly labeled. Please report the tumor content (of the labeled tumor area).
MSI only: Normal tissue in addition to tumor tissue:
- 1–2 ml EDTA blood (recommended sample type) or
- 1–2 µg DNA or
- FFPE block with normal tissue of the patient
- If possible: H&E-stained slides with tumor and (if a blood sample is not available) normal tissue area distinctly labeled. Please report the tumor content (of the labeled tumor area).
Here you can find more information on how to ship your sample safely.
Turnaround Time
- Turnaround Time: 2–3 Weeks
Costs
The prices for our human genetic diagnostics depend on the size of the selected Diagnostic Panel and the selected gene sets. All prices include sequencing, bioinformatic analysis, and issuing of a medical report by our team of experts in human genetic diagnostics.
This Is What Makes Our CancerEssential® Service Special
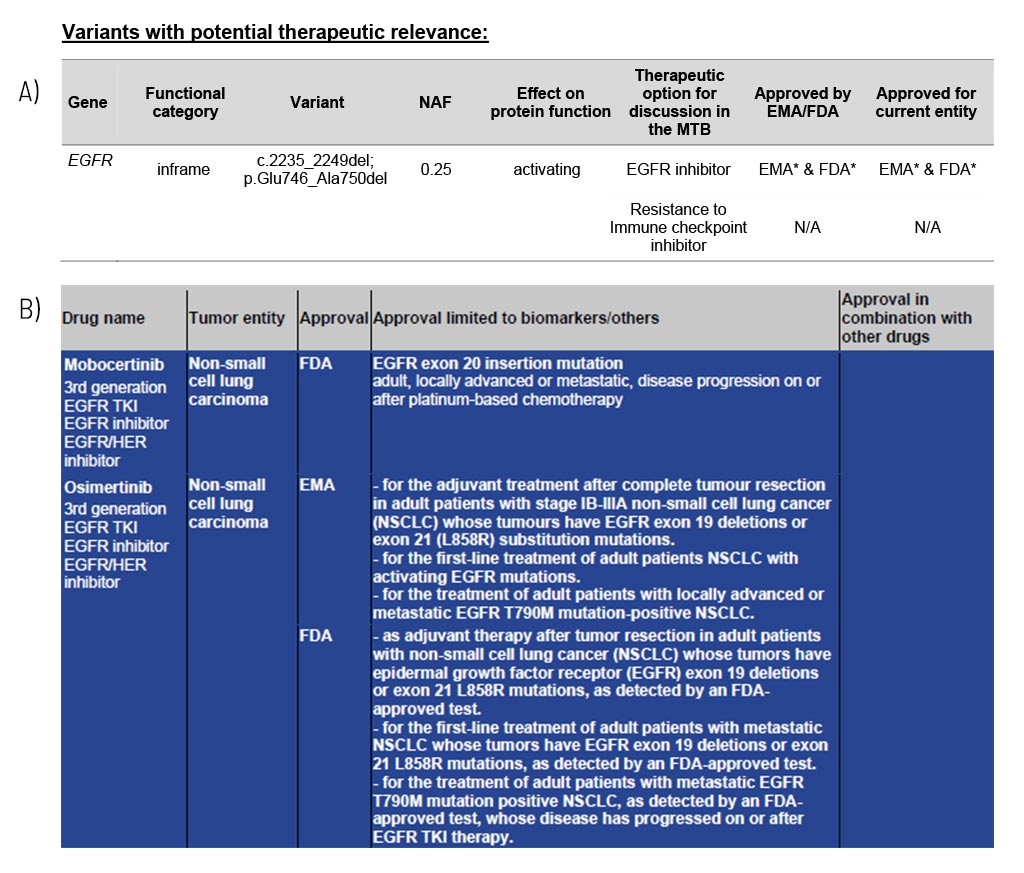
Variants with Potential Therapeutic Relevance
Guidance on potentially effective drugs
For each gene, the somatic change is depicted in detail, and the resulting therapeutic options are stated, including the EMA/FDA approval (A). These options are the basis for discussion in a molecular tumor board (MTB).
At the end of the medical report, in the appendix/supplement, we provide an extensive list of possible therapeutic strategies for each identified somatic change (B). This list includes drug classes and names as well as their approval (FDA/EMA) and limiting conditions.
Sample Report: Exemplary for the EGFR variant detected and the resulting therapeutic options in a patient with non-small cell lung cancer. Top panel (A): An excerpt from Table 1 of the findings, listing variants with therapeutic relevance. Lower part (B): An excerpt of the drug listing. In addition to the drugs shown, other drugs are also described.
Structural Variants (9 Genes)
Selected therapy relevant fusion are evaluated additionally without further costs.
NTRK1, FGFR1, FGFR2, FGFR3, BRAF, ALK, RET, MET, ROS1
Contact Us
Do you have a question, or are you interested in our service?
Diagnostic Support
We will assist you in selecting the diagnostic strategy – for each patient.