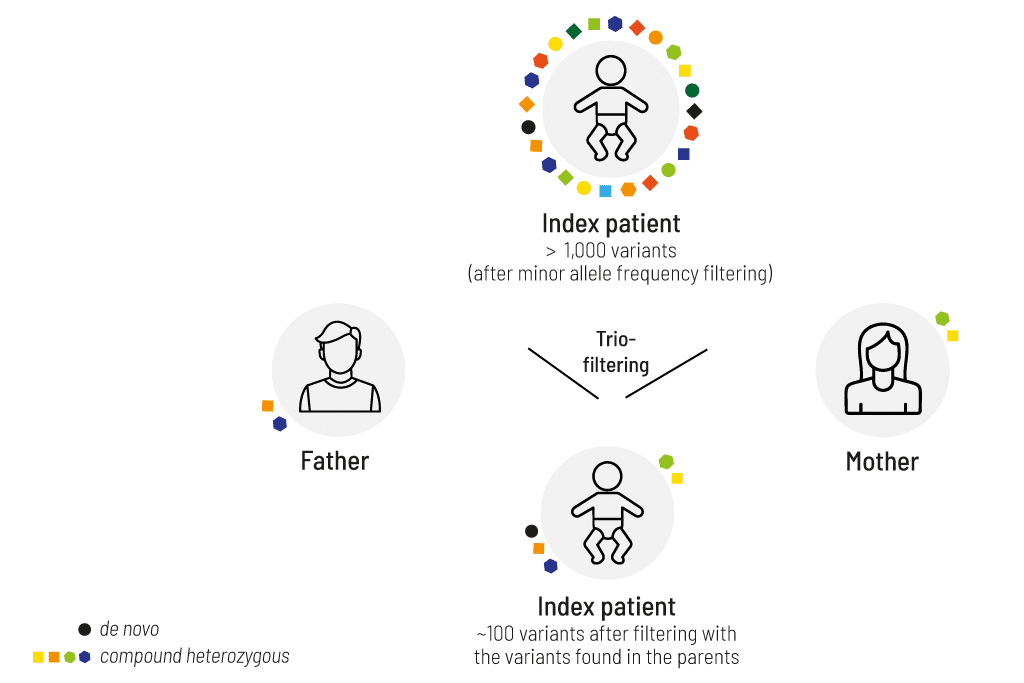
Trio ExomeXtra® is used to diagnose an affected patient with unaffected parents. Including both unaffected parents in the analysis significantly increases the chances of a successful diagnosis. For Trio ExomeXtra®, the exomes of the parents and the index patient are sequenced. This allows an exome-wide segregation analysis and, thus the highest solution rates. Comparative exome diagnostics is particularly effective because the number of variants to be evaluated is minimized, and single variants’ numerous and cost-intensive segregation analyses are avoided. In addition to SNVs and CNVs, we also check for uniparental disomies (UPD), covering both isodisomies as well as heterodisomies affecting whole or partial chromosomes.
Are you insured in Germany? Our colleagues at the Zentrum für Humangenetik Tübingen will gladly support you!
What We Offer with This Service
Our Promise to You
Service Details
Medical Report with:
- variants with clinical relevance (ACMG class 3, 4, 5) — more information
- interpretation & discussion of these variants and the affected genes
- interpretation of genetic relevance for family planning
- recommendations for clinical disease management & further tests
- genome-wide detection of copy number variants (CNVs) — more information
Additional Services:
- ACMG gene panel — more information
- pharmacogenetics — more information
- HLA typing — more information
Sample Report
Our Standard Sample Requirements
- 1 ml–2 ml EDTA blood (recommended sample type)
- Genomic DNA (1 µg–2 µg)
- DBS cards, buccal swabs, or saliva are also possible
Here you can find more information on how to ship your sample safely.
Other sample material sources are possible on request. Please note: In case of insufficient sample quality, the analysis might fail. If you have more than one option of samples, please contact us (diagnostic-support@cegat.de) and we will assist you in choosing the optimal sample for your patient.
Extra Insightful Results
In the evaluation process, our experienced team investigates all variants according to the latest scientific knowledge. For variants in genes that are not clearly associated, with the patient’s suspected phenotype, we predict the possible contribution through extensive additional literature research. If we find evidence that a variant not yet described contributes to the patient’s phenotype, this variant is described in our medical report. Comparison of the affected patient’s data with those of the unaffected parents (figure 1) allows us to identify the following SNV/CNV combinations:
- de novo — new in the index, not present in parents
- homozygous — homozygous in the index, heterozygous in both parents
- compound heterozygous — two or more variants in the same gene in the index on different alleles; each parent is a heterozygous carrier of one variant
- X-linked — male patient is hemizygous, the mother is heterozygous
- loss of heterozygosity — index is homozygous, one parent heterozygous, other parent wildtype
- parental mosaicism — index is heterozygous, one parent has a genetic mosaicism
Benefit From Our Comparative ExomeXtra®
Our Unique Analysis Strategy Considers Often Overlooked Issues, Such as Mildly Affected Parents
In comparative family exome diagnostics, not only the affected patient but also other relatives are sequenced. The most commonly performed analysis is the trio exome: the inclusion of both unaffected parents of the index patient highly increases the chances of diagnostic success to be approximately twice as high compared to single exome diagnostics.1
In family constellations where it is not possible to obtain a sample from both genetic parents, or one parent might be affected as well, the adaptive ExomeXtra® bioinformatic pipeline allows us to analyze all kinds of family combinations, like duos. This goes beyond commonly performed analyses of private and shared variants between two persons.
Our additional analyses are the best approach for solving cases with only one parent (affected or unaffected) or any other indexrelative constellations. A detailed phenotypic description of all affected individuals is the basis for a precise and successful data interpretation. Standard trio exome diagnostics rely on the assumption that both parents are not affected. This filtering ignores that some parents are only mildly affected, for example, which would result in negative findings. We compensate for these situations with a unique analysis strategy that allows us to solve cases where the phenotype is caused by variants with:
- reduced penetrance
- variable expressivity
- imprinting effects
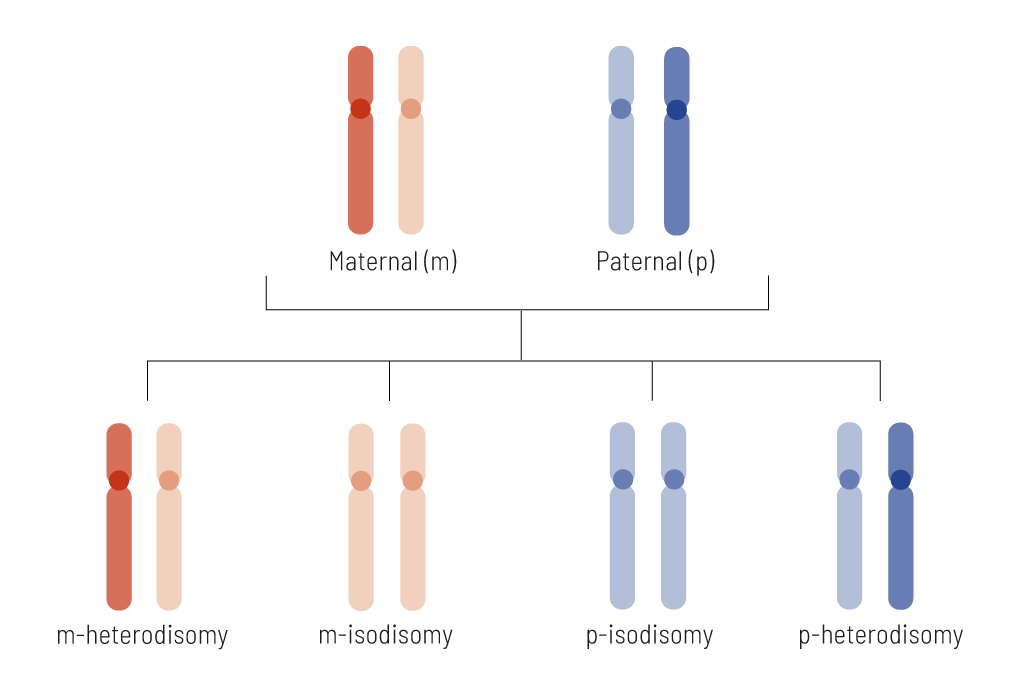
Detection of Uniparental Disomies (UPDs)
We also detect and report uniparental disomies (UPDs) in trios. UPDs can cause diseases due to imprinting effects, underlying homozygous pathogenic variants, or low-level mosaic aneuploidies.
We are able to report on all four possible UPD constellations: maternal heterodisomies maternal isodisomies, paternal heterodisomies, and paternal isodisomies (figure 2). We cover UPDs of entire chromosomes, as well as segmental ones.
Figure 2: Schematic illustration of the inheritance pattern of uniparental disomy (UPD). UPD occurs when both pairs of chromosomes are inherited from only one parent, which means the other parent’s chromosome for that pair is missing. The figure shows the possible types of UPDs that can occur, which may be inherited maternally (both red) or paternally (both blue). UPDs are also further sub-categorized depending on the constellation; if one chromosome is inherited in a duplicated fashion (isodisomy) or if each chromosomes in the pair is inherited (heterodisomy).
References
1 Farwell, K. D. et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genetics in medicine : official journal of the American College of Medical Genetics 17, 578–586; 10.1038/gim.2014.154 (2015).
Further Information
Webinar: Learn How We Can Help You Solve Complex Patient Cases
Webinar: Better than exome, smarter than genome
Dr. Dr. Saskia Biskup about ExomeXtra®
Trio exome diagnostics: One of the most powerful tools in genetic diagnostics
Designing the best possible exome
Identifiy the disease-causing alteration in a fetus
Exome diagnostics saved a little girl’s life
Downloads
Contact Us
Do you have a question, or are you interested in our service?
Diagnostic Support
We will assist you in selecting the diagnostic strategy – for each patient.