Recent research suggests a close relationship between the composition of the human microbiome and the occurrence of a variety of diseases, including the response towards pharmaceutical drugs. According to recent findings, this relationship has been particularly shown for the gut microbiome, but it can also be extended to other body sites, such as the skin, mouth, or nose. Furthermore, microorganisms are involved in many biochemical reactions in the environment and represent important constituents of environmental ecosystems. Understanding microbial functions in specific microbiomes or host-microbe relationships, offers great potential for new therapeutic discoveries, especially for microbes, which are difficult to cultivate.
Shotgun metagenomic sequencing analyzes the complete DNA content of a sample and allows accurate detection of microbes (bacteria, archaea, fungi, protozoa, viruses, etc.) down to species level. Accordingly, functional genes encoding specific metabolic enzymes can be analyzed. Shotgun metagenomic sequencing is the best choice when microbiomes need to be thoroughly characterized, including accurate identification of microbial species and their functional repertoire.
Applications of shotgun metagenomic sequencing are diverse and include:
- disease monitoring
- microbial biomarker detection
- drug development
- characterization of environmental microbiomes
- discovery of new microbial species (de novo assembly)
We offer different Shotgun Metagenomic Sequencing products to address a variety of research questions.
CeGaT Is the Best Partner for Sequencing Your Project
Our Commitment to You
Fast Processing
Turnaround time
≤ 15 business days
High Quality
Highest accuracy for all processes
Secure Delivery
Secure provision of sequenced data via in-house servers
Safe Storage
Safe storage of samples and data after project completion
Our Service
We provide a comprehensive and first-class project support – from selecting the appropriate product to evaluating the data. Each project is supervised by a committed scientist. You will have a contact person throughout the whole project.
Our service includes:
- detailed project consulting
- product selection tailored to your project
- detailed bioinformatic evaluation of your data
- detailed project report with information about sample quality, sequencing parameters, bioinformatic analysis, and results
Benefit from our dedicated support and accredited workflows.
Explore Our Product Portfolio for Shotgun Metagenomic Sequencing
We offer different products for shotgun metagenomic sequencing to address a variety of research questions. Would you like to have bioinformatic analyses performed on your data in addition to the included deliverables? Each of our products can be supplemented with further services. We are happy to advise you.
Shotgun Metagenomics Classic | Shotgun Metagenomics Flex |
Species | Species |
Sample types | Sample types |
Target | Target |
PCR amplification | PCR amplification |
Read length | Read length |
Sequencing platform | Sequencing platform |
Output | Output |
Included deliverables | Included deliverables |
Bioinformatics
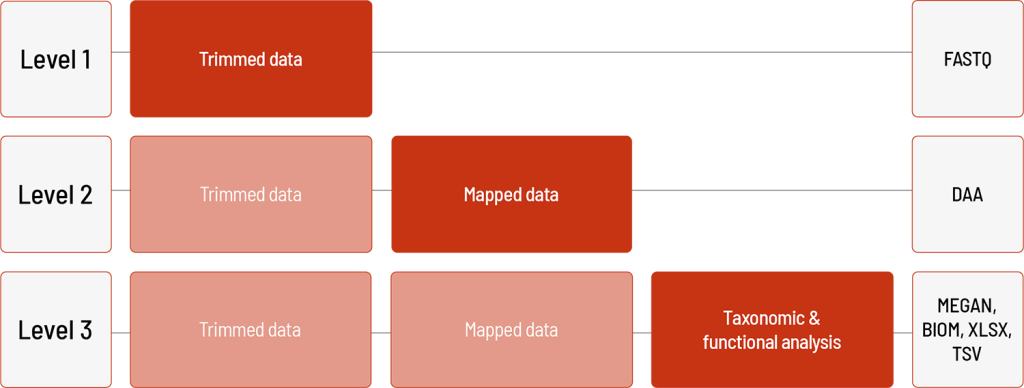
Raw sequencing data are automatically processed. We offer different levels of bioinformatic analysis. The default level is Level 1. With increasing bioinformatic level, more data are delivered. All higher levels include the data from the lower levels. In addition to the data, and independent of the analysis level, a project report is generated.
Level 1:
- demultiplexing and adapter trimming of the sequencing data (FASTQ format)
Level 2:
- mapping of the sequencing data against microbiome database (DAA format*)
Level 3:
- taxonomic and functional classification using MEGAN 6 (MEGAN format*)
- generation of taxonomic abundance tables including sequence counts and relative abundances down to the species level (XLSX and BIOM format)
- generation of microbial function tables including sequence counts and relative abundances of functional genes classified using the KEGG database (XLSX format)
- generation of bar plots showing relative abundances down to the species level (PNG format)
* These files can be interactively explored and further analyzed using MEGAN6, a user-friendly and open source software for analyzing microbiome data.
Technical Information
At CeGaT, paired-end sequencing (2 x 100 bp) is performed using the Illumina sequencing platforms. If you require other sequencing parameters, please let us know! We can provide further solutions.
We are pleased to offer you our specifically designed stool sampling kit, which provides convenient and unbiased self-collection of fecal material combined with our DNA isolation service. Please also reach out to us to discuss sampling and DNA extraction of various starting material.
Further Information about Shotgun Metagenomic Sequencing
With the shotgun metagenome analysis, the whole genome of all organisms in a sample are sequenced. This contrasts with the 16S sequencing, where only one gene is sequenced. Thus, shotgun metagenomic sequencing is comparable to whole genome sequencing. However, not one but many organisms are sequenced simultaneously. The advantage of shotgun metagenomic sequencing is its versatility, as one is not restricted to bacteria and archaea, but can also identify eukaryotes and viruses. The area of applications ranges from the analysis of the gut microbiome, over other body sites, such as the oral or nasal cavity, to environmental samples, e.g., soil samples. With shotgun metagenomics, all organisms of a sample can be taxonomically differentiated down to the species level. Additionally, a functional analysis is possible with shotgun metagenomic sequencing. For a sufficient amount of reads, shotgun metagenomic sequencing is more powerful to identify less abundant taxa in comparison to 16S sequencing.
For shotgun metagenomic sequencing, the DNA is extracted from all cells in the sample. This DNA is then fragmented into small pieces that are independently sequenced. In contrast to 16S sequencing, no specific genetic locus is required as target. All sequenced fragments can be aligned to loci of different genomes in the sample. These different genomes are not restricted to bacterial genomes. For the alignment of the sequenced fragments, the 16S rRNA sequence and biologically informative sequences are used.
Shotgun metagenomic analysis can also be used to measure the taxonomic diversity of a sample.
To determine the taxonomic diversity, different approaches are available.
- In marker gene analysis, a sequence is compared to a database of known taxonomically informative sequences. These taxonomically informative sequences are also called marker genes. If the sequence is a homologue of the marker gene, an algorithm classifies and annotates similarity of the sequence.
- Another approach is called binning, where sequences are clustered into groups. In the compositional binning method, the metagenomic sequences are clustered into groups. The grouping is based on their composition. In the similarity binning, the sequences are clustered based on their similarity to the taxonomically annotated sequences. The fragment recruitment binning method aligns sequences to other, almost identical, sequences. This binning method results in a metagenomic estimate of the genome.
By elucidating the taxonomic diversity of a sample using shotgun metagenomic sequencing, microbiomes can be thoroughly characterized. Thus, shotgun metagenomic sequencing can help monitoring diseases associated with an altered microbiome, detecting microbial biomarkers, developing drugs, characterizing environmental microbiomes, and discovering new microbial species. Shotgun metagenomic sequencing increases the understanding for the functions and characterization of specific strains, leading to therapeutic discoveries and innovative ways to synthesize novel products.
Downloads
Contact Us
Do you have a question or are you interested in our service? Feel free to contact us. We will take care of your request as soon as possible.
Start Your Project with Us
We are happy to discuss sequencing options and to find a solution specifically tailored to your clinical study or research project.
When getting in contact, please specify sample information including starting material, number of samples, preferred library preparation option, preferred sequencing depth and required bioinformatic analysis level, if possible.