The introduction of next-generation sequencing (NGS) fundamentally changed how we approach scientific and diagnostic issues. Next-generation sequencing’s potential to produce large amounts of sequence data for a relatively low price led to a series of groundbreaking discoveries and ushered the field of genomics into a new era. Third-generation sequencing further has revolutionized the detection of genetic mutations owing to the single-molecule real-time (SMRT) technology.
The combination of five Illumina NovaSeq™ 6000 and two NovaSeq™ X Plus sequencing systems for high-throughput sequencing and PacBio’s Sequell IIe for high-quality long-read sequencing, provides a wide spectrum of sequencing applications for research projects and pharma studies.
CeGaT has specialized in the use of NGS since its foundation. We continuously invest in the newest technologies to aim for the most accurate and reliable results for our customers. Meanwhile we can offer our customers third-generation sequencing (TGS) on the PacBio sequencing platform. Our profound diagnostic knowledge and bioinformatic expertise complements the state-of-the-art lab platforms and sequencing technology. We offer the knowledge, the latest technology and award-winning service for your projects.
Our team has a substantial experience with human and other mammalian samples and a wide range of expertise in the processing of plant samples in addition to microbial samples.
Based on many years of using NGS technology, we have gained great expertise in high-throughput technology for processing thousands of samples. Our laboratories are equipped with liquid-handling workstation platforms from Hamilton. These platforms can be individually programmed allowing specific adaptations of our protocols based on your requirements.
The multiplexing strategy we have established in our laboratory allows the sequencing of hundreds of samples in one sequencing run, or even in one lane, and therefore represents true high-throughput sequencing.
We would be happy to demonstrate the feasibility of your high-throughput projects in a pilot study.
Illumina Sequencing Platforms
Illumina NovaSeq™ X Plus System
- For high-throughput sequencing with exceptional accuracy
- Ideal to perform larger studies and more data-intensive projects
- Produces up to 16 Tb data in 48 hours per dual flow cell run
Find a performance comparison of our NovaSeq™ devices here.

Illumina NovaSeq™ 6000 System
- For high-throughput short-read sequencing with lowest cost-per-base
- Ideal for large-scale and deep-sequencing projects
- Produces up to 6 Tb data in 20 hours
Find a performance comparison of our NovaSeq™ devices here.

Illumina MiSeq™ System
- For smaller sequencing projects with multiple configurations
- Read length up to 2 x 300 bp

Illumina Workflow
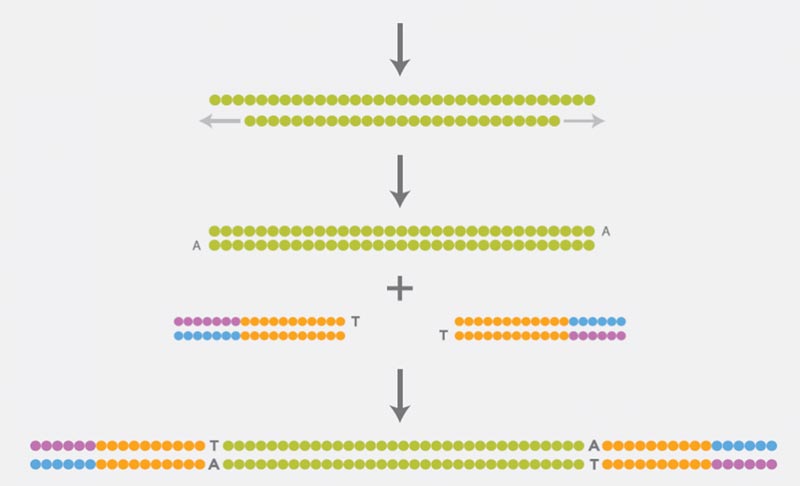
Preparation of a DNA Library
For the preparation of the DNA library, the DNA must first be fragmented. The ends of the DNA fragments are then repaired, as they are damaged by the fragmentation. The ligation of adapters to both sides of the repaired DNA fragments completes the preparation of the DNA library. These adapters contain sequence motifs that are required for subsequent steps (clonal amplification and the actual sequencing).
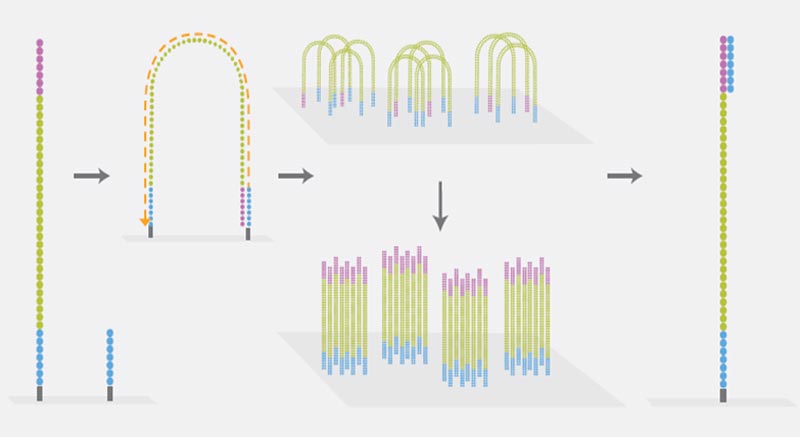
Clonal Amplification
Illumina sequencing technology uses the so-called Bridge-PCR for clonal amplification of individual DNA fragments. This PCR is performed on the flow cell (a glass side similar to a microscope slide) on which the actual sequencing takes place. DNA fragments are linked via the adapters to the flow cell, then the Bridge-PCR is carried out. Oligos bound to the flow cell function as primers, and each individual DNA fragment forms a separate cluster of identical DNA fragments.
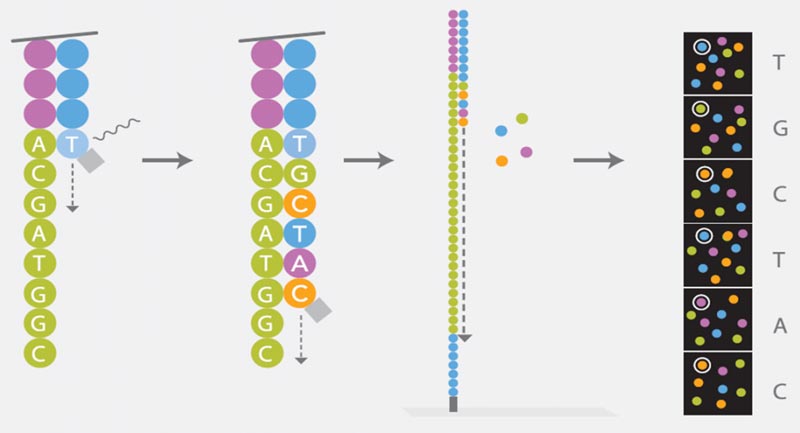
Sequencing
Illumina’s Sequencing by Synthesis (SBS) is based on the Cyclic Reversible Termination (CRT) method. Each of the four nucleotides is linked with a different dye and modified by a terminator group. During a reaction cycle, all four nucleotides are offered simultaneously to the polymerase for strand synthesis. After incorporation of a complementary nucleotide strand, elongation is no longer possible due to the blocking effect of the terminator group. The four dyes of each nucleotide can be detected using Imaging: the dye and the terminator group are cleaved and a new synthesis cycle starts. The sequence of each cluster is thus simultaneously determined base by base.
PacBio Platform
PacBio Revio System
- For high-quality long-read HiFi sequencing
- Sequence up to 40 human genomes per week

PacBio Sequel® IIe System
- For high-quality long-read HiFi sequencing
- Sequencing projects with reads up to 25 kb and 99,9% accuracy

PacBio Workflow
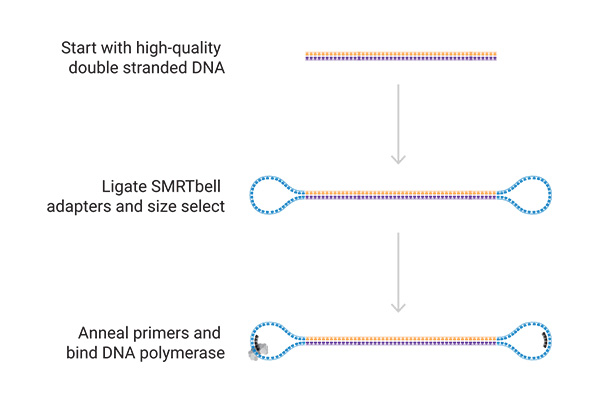
Preparation of a Library
- For high-quality long-read HiFi sequencing
- Sequencing projects with reads up to 25 kb and 99,9% accuracy
Sequencing
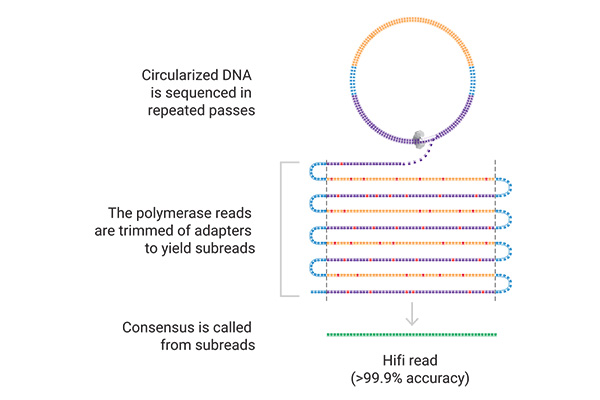
On the sequencer, the SMRT® cell contains millions of wells called zero-mode waveguides (ZMWs). Single DNA molecules are immobilized in these wells, and the polymerase incorporates labeled nucleotides.
In this process, light is emitted, and nucleotide incorporation is measured in real-time. The reactions are recorded and can be analyzed. When sequencing in circular consensus mode (CCS), highly accurate long reads (HiFi) are generated because SMRT sequencing can sequence the same DNA molecule multiple times.
10x Genomics® Platform
10x Genomics® Chromium™ X Controller
- For analysis of single cells at a massive scale
- Microfluidics system to capture tens of thousands single cells per sample

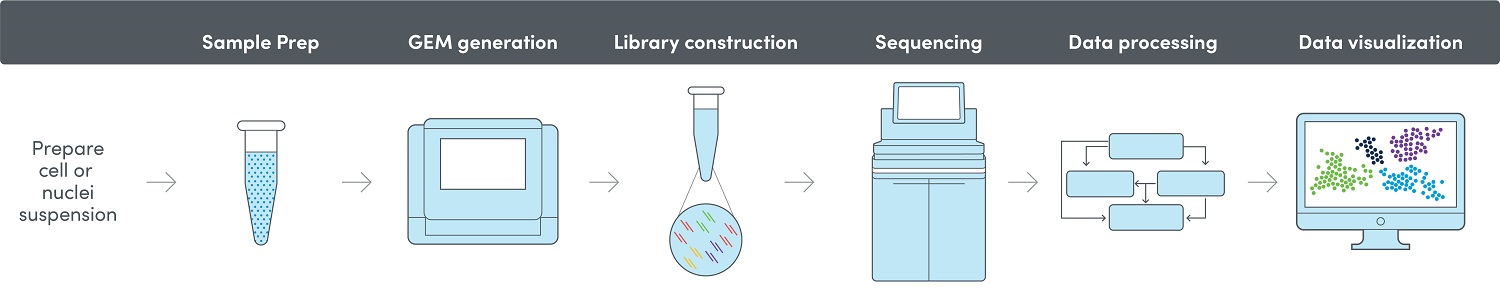
10x Genomics® Workflow
Generation of Gel Beads in Emulsion
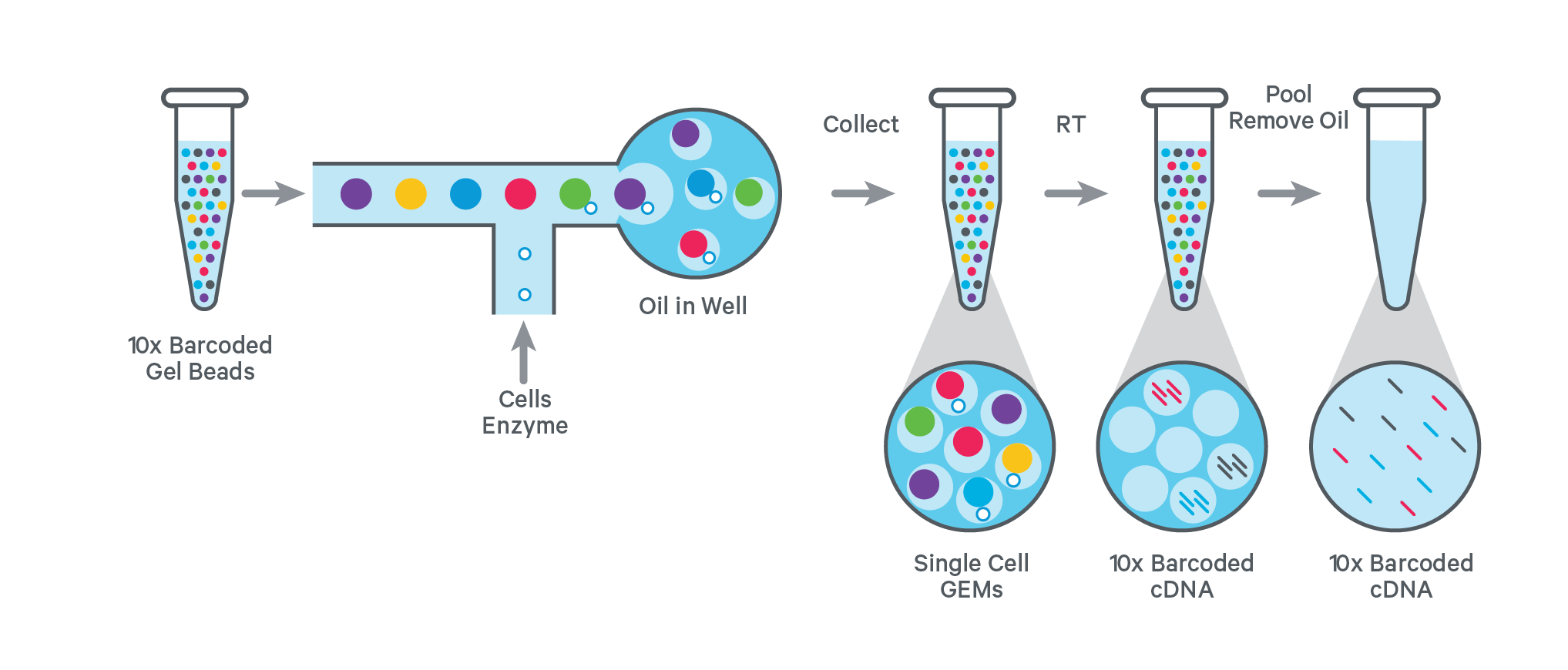
The workflow for generating a single-cell library starts with combining single cell suspensions with reagent mix and 10x barcoded gel beads. On the chromium chip, gel beads in emulsions (GEMs) are generated by combining cells and gel beads in a single aqueous stream in a channel within the chip. Droplets are formed while passing the recovery oil interface and are collected. Most of the collected GEMs contain a gel bead and a subset includes a single cell.
Preperation of a Library and Sequencing
After GEM generation, the gel beads dissolve and release oligos and enzymes into the solution to enable the amplification. This results in many copies of one cell with the same barcode but a unique molecular identifier (UMI).
After barcoding, the barcoded fragments are pooled together, and the final library construction is performed in bulk. Sequencing of the libraries is conducted on the Illumina NovaSeq™ 6000 or MiSeq sequencing system using the 10x.
Contact Us
Do you have a question or are you interested in our service? Feel free to contact us. We will take care of your request as soon as possible.
Start Your Project with Us
We are happy to discuss sequencing options and to find a solution specifically tailored to your clinical study or research project.
When getting in contact, please specify sample information including starting material, number of samples, preferred library preparation option, preferred sequencing depth and required bioinformatic analysis level, if possible.